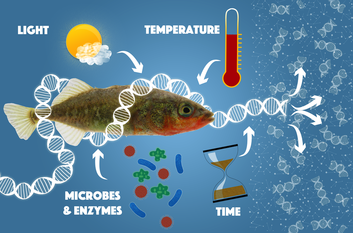
 This summer I had the awesome opportunity to go to Wales to attend an Environmental DNA (eDNA) conference and workshop at Bangor University. To be honest, I had no idea what eDNA was, why anyone should care, or even why I was going before I started my program (but I was going first class so did it even matter?). But I am really glad I did because eDNA is one of the most exciting new scientific method for studying biology and the environment and I hope to use it during my PhD. So what is eDNA? What is that Jay? Well eDNA is officially any piece of DNA that can be amplified directly from the environment, essentially water, air, soil, ice, and blood counts too for some reason. Apparently when organisms move around through the water or soil they leave behind hundreds of cells (maybe free pieces of DNA?), which end up being free floating in the environment (next time I swallow sea water I will be reminded of all the lovely pieces of eDNA and Enterococcus I am eating). Scientists can the extract the DNA from these free floating cells collected from the environment, just like they would any other animal. What makes this so revolutionary, is that now a scientist walk into a river or stream with a glorified bucket, extract, amplify, and sequence the DNA and boom (after lots of hours of bioinformatics work): we know which animals, microbes, plants, you name it are living in there. Of course we need to already have DNA sequences of all the organisms that do live there, and we have to do a lot more testing to figure out how long eDNA can last in the environment and how much eDNA comes off of different organisms, but the potential for eDNA to revolutionize ecology and environmental science is there and the technology to do this is only going to get better and better (unless of course we get a Walking Dead Scenario or El Niño washes California away). Current methods for surveying biodiversity are fun. But they are REALLY time intensive, and expensive, and they require a ton of taxonomic expertise, and they are hard to repeat. eDNA offers a rapid and affordable method for evaluating biodiversity. (Currently it costs about ~$200 an eDNA sample, but as sequencing technology becomes cheaper and cheaper, this price is only going to go down) Thus eDNA is an especially important method for looking at marine and aquatic ecosystems, because now we have the tools to rapidly and affordably survey the entire biodiversity of an area. Now we can relatively easily compare marine biodiversity of dozens if not hundreds of sites to look at patterns of biodiversity and what forces shape them, which has been a long time interest of my lab. And for me personally, I think that the greatest potential for this technique is to observe how human impacts are causing changes in marine biodiversity and ecosystem function. With eDNA, I hope to investigate the synergistic effects of global (ocean acidification and climate change) and local (overfishing, habitat degradation, pollution) human impacts on marine ecosystems. We know quite a bit how overfishing can wreck an ecosystem, that ocean acidification is threatening to make large parts of the ocean uninhabitable for calcifying organisms, and that nutrient runoff from over-fertilized farms and sewage treatment plants can lead to massive coastal dead zones. But what happens when you add all three into the pot? (My guess is that is isn't good). Still, this is incredibly important information that coastal managers need to know to protect our marine ecosystems into the future. So eDNA looks like the metagenomic tool that marine biologists have been looking for. I hope to use eDNA to rapidly assess fish and microbe biodiversity of the Channel Islands and Santa Monica Bay during and after this Chuck Niño to look at how marine protected area design affects ecosystem resilience and what California's marine ecosystems might look like under further climate change (Yellow bellied sea snakes?!?). Plus it's potentially an amazing opportunity for citizen science as almost anyone can collect a bucket of seawater. And one day in the distant future little robots might swim the seas, collecting eDNA and sequencing it, pinging back the entire biodiversity of the ocean every couple of hours. Then we can finally find where the Grunion go and where the giant squid like to party. I can't wait.
1 Comment
11/7/2024 05:44:33 am
Wow, eDNA sounds like a game-changer for environmental science so exciting to see how this innovative method can revolutionize our understanding of marine ecosystems and biodiversity!
Reply
Leave a Reply. |
AuthorZack Gold Archives
June 2019
Categories |
 RSS Feed
RSS Feed